This morning, I stumbled across a fantastic website, called Molecular Movies. It describes itself as a 'portal to cell and molecular animation' and aims to provide scientists with tutorials on developing 3D visualisation skills.
Alongside tutorials, the website has a showcase section, with examples of 3D being used to visualise complex biological scenes - although these are interesting to watch, the scientific content is a bit beyond my own knowledge. Fortunately (and more importantly), it is useful to have a 'database' of the kind of work that is going on in my field.
Returning to the learning resources, I realised I had already covered the majority of content, as it was designed for those new to using 3D. That is, until I found an interesting article on using metaballs to create molecular surfaces, using Cinema 4D. Although I was not familiar with using C4D, I decided it was worth looking at it, as it was going to less technically challenging than learning Houdini (which is also reliant on Python), and should give a similar result.
Creating metaballs in C4D was fairly straight forward, and gave great results with very little input. It took a bit of time to figure out how to animate objects, and then render a scene, but after using the software, I would definately be confident using it again. The geometry was also significantly 'cheaper' than my testing in RealFlow, and could be used on a large scale. A video example of metaballs in action can be seen below;
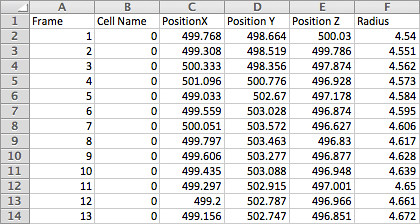
The next step was to take my cell data-set and use metaballs to create a single organic structure - this proved substantially more difficult. Realising that I couldn't simply 'read' my data, as C4D also relies on Python, I opted to export both OBJ and FBX files from Maya, hoping that at least one would work.
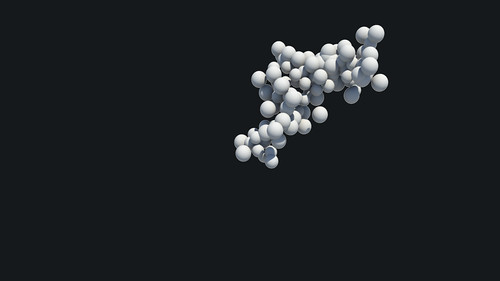
These imported easily, and a metaball surface could be applied, but did not work properly (simply creating one large sphere). After vast amounts of experimentation, I scaled the imported objects... and success! A simple fix for what seemed like a complicated problem. This breakthrough meant that I could now take objects into Cinema 4D, use metaballs to create a surface, and either render, or export to Maya for render (as I am more familiar with the software package).
The only difficulty now, is that this workflow currently only works on single frames, and can't be applied to my animated data-set... this is the next problem to solve! A render of the results so far can be seen below (showing a before/after comparison);
After my previous post, I felt like I had reached a brick wall, but after discovering the Molecular Movies website, it has helped me hurdle these difficulties... only to find a new hurdle waiting on the other side!